diversity sets

B. oleracea diversity - photo courtesty WHRI 2002 (R.Sampson)
A Diversity Fixed Foundation Set (DFFS): is defined as
"an informative set of genetically fixed lines representing a
structured sampling of diversity across a genepool"
| Species | Set | Number of lines | Founder line seed available |
Fixed line seed available | Founder DNA available | Fixed line DNA available | detail | MTA | Project | Contact |
|---|---|---|---|---|---|---|---|---|---|---|
| B. napus | BnaDFFS | 189 | 152 | 57 | 152 | 53 | - | Graham Teakle (WHRI) |
||
| B. oleracea | BolDFFS | 376 | - | - | 370 | - | - |  |
Graham Teakle (WHRI) |
|
| B. rapa | BraDFFS | - | - | - | - | - | - | - | - | Guusje Bonnema (WUR) |
| B. C genome | BCgDFS | 88 | - | - | 88 | - | - |  |
Paul Hand (WHRI) |
 A web based User Inteface has been developed to query the population Area of CropStore, to view data summaries and download Excel workbooks containing detailed data - Available Here (CropStore :: Interface)
A web based User Inteface has been developed to query the population Area of CropStore, to view data summaries and download Excel workbooks containing detailed data - Available Here (CropStore :: Interface)More information
- Background and concept
- Publication describing generation of fixed lines (Pink et al., 2008)
- Availability and distribution
- Data collation
- Current and planned status
Development of Diversity Sets for long-term community-wide molecular and trait exploration of Brassica gene-pools
Graham King, Graham Teakle, Dave Astley, Dave Pink. WHRI, March 2004Modern crops contain a surprisingly narrow genetic base, despite the existence of extensive allelic variation within germplasm collections. Although breeders exploit the existing natural variation for particular target traits within crop species, there has been considerable reduction in biodiversity through canalisation of alleles. Introduction Core Collections have been developed for Brassica spp in the past, within EU GenRes and other programmes. These were based on accessions held within public genetic resource collections (see 1; 2; 3;) . Seed were multiplied and accessions screened for a number of fungal and other resistance traits. Whilst valuable in identifying 'hotspots' of variation within the relevant gene-pools, these collections consisted of heterogeneous and heterozygous materlal. This limits their long-term use for correlating detailed genetic studies.
The Concept
 In the course of recent molecular analysis we have found considerable genotypic variation within individual extant genetic resource accessions. This is present both as within-line heterogeneity and within-plant heterozygosity. As such, any co-ordinated trait and genetic studies will be very difficult to interpret unless material with acceptable levels of homozygosity is produced and made publicly available.
To address this issue and provide researchers with relevant resources, 'Diversity Fixed Foundation Sets' (DFFS) will be constructed.
These are designed to represent the majority of allelic variation within respective gene pools in a form suitable for long term exploitation by researchers and end users.
Theoretical studies (Lawrence MJ, et al (1995) Euphytica 82:89-99; 101-107) indicate that 400 homozygous accessions, collected from throughout the world, should contain 99% of the allelic polymorphism (i.e for alleles with frequencies >2%) present in the species.
see also 4
In the course of recent molecular analysis we have found considerable genotypic variation within individual extant genetic resource accessions. This is present both as within-line heterogeneity and within-plant heterozygosity. As such, any co-ordinated trait and genetic studies will be very difficult to interpret unless material with acceptable levels of homozygosity is produced and made publicly available.
To address this issue and provide researchers with relevant resources, 'Diversity Fixed Foundation Sets' (DFFS) will be constructed.
These are designed to represent the majority of allelic variation within respective gene pools in a form suitable for long term exploitation by researchers and end users.
Theoretical studies (Lawrence MJ, et al (1995) Euphytica 82:89-99; 101-107) indicate that 400 homozygous accessions, collected from throughout the world, should contain 99% of the allelic polymorphism (i.e for alleles with frequencies >2%) present in the species.
see also 4DFFS Stucture
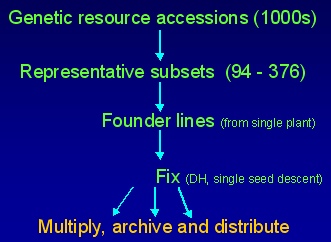
WHRI first initiated the establishment of a DFFS for B. oleracea, which is comprised of a structured sample of 386 accessions representing the diversity within the gene-pool. These have been selected as nested subsets of 94 (so DNA samples fit on a 96-well microtitre plate, with 2 controls). Each subset has been selected to represent diversity within species subtaxa and crop type, as well as variation in eco-geographic origin. The sets are being fixed through generation of doubled haploids and inbreeding to establish 'immortal' lines which may be used by a wide range of researchers and breeders for trait and molecular assessment. The approach of generating fixed lines is particularly powerful as the data obtained are cumulative, allowing long-term comparative analysis. In addition, fixed lines enable experimental trials to be established with replicate plants. As such they may be interpreted to provide insights into contribution and interaction of genetic, environmental and developmental components of variation. Details of the composition of the founder sets will be presented in the near future.Development of the sets
The development Diversity Fixed Foundation Sets involves the following steps:
- Survey available genetic resources and related information
- Identify and source accessions to represent diversity
- Grow sample of plants and select representative 'founder plant'
- Extract DNA of founder plant - prepare GenomiPhiTM sample
- Bulk seed of founder plant
- Preliminary assessment of variation in founder line
- Fix founder line by microspore (doubled haploids) or single seed descent selfing (inbred line)
- Extract DNA from fixed lines and make available
- Bulk up seed of fixed lines and make available
Uses
- Graphical Genotyping using mapped markers
- Allelic variation in candidate genes
- Phenotypic trait assessment
- Association genetics and Linkage disequilibrium
Availability and distribution of the DFFSs
For B. napus, B. oleracea, and Brassica C genome Diversity Sets - from University of Warwick (WHRI)Seed
The University of Warwick is committed to distributing seed for the DFFSs under the standard Materials Transfer Agreement (sMTA) that accompanies the International Treaty on Plant Genetic Resources for Food and AgricultureThe DFFSs require considerable cost and effort to develop. If the use of a well characterised, structured sample of the genetic diversity in a genepool is integral to your research, we suggest an appropriate acknowledgement of the work done to develop the DFFS may be to include the person responsible at the University of Warwick as a co-author on the first paper describing the use of the DFFS. By this we are not intending to attach any strings and if we have no further involvement in future publications then an acknowledgement/reference to the original paper would be sufficient. We would of course very much welcome collaborative follow up research.
Distribution is subject to availability and, for the fixed lines, will depend on progress through the fixing process. The current status of availability will either be listed in the Excel workbook files describing the DFFSs (see top of this page) or can be obtained by contacting Graham Teakle.
DNA
DNA is available for most of the DFFS founder lines and will be for the fixed lines in due course. This will be GenomiPhiTM amplified DNA in the range of 100-500 ng.- A small handling charge will be charged for requests of DNA - please contact Graham Teakle for details.
Charges
Unfortunately, due to the considerable cost in generating seed for these populations, we have to make a charge to cover the cost of replacement of the seed in order to ensure future availability. These charges are currently:- £20 per line for self-compatible populations (BnaDFFS) for starter pack of seed (approx. 0.5g)
- £50 per line for self-incompatible populations for a starter pack of seed (5-25 seed, depending on availability)
- Recipients are then free to regenerate further seed to meet their future requirements.
Data collation
WHRI will collate genotype and phenotypic data into a structured database (a module of the CropStore family of databases), and make available over the web through online interfaces, XML etc. The genotype data will become integrated into other online Brassica genome and genetic databases.Current status
- BolDFFS: B. oleracea DFFS. We anticipate generating 4 x 94 = 376 lines - for details see table above
- BCgDFFS: C genome DFFS. We anticipate generating 94 lines of C genome diploid species - these are n=9 and able to inter-cross with B. oleracea. Species include B. cretica, B. incana, B. macrocarpa, B. villosa et al. More details will be made available in the near future.
- BnaDFFS: B. napus DFFS is being developed initially through the Defra OREGIN project. We are very keen to ensure that a wider representation of the gene-pool is represented and are working with partners worldwide to ensure that this DFFS is scalable. We anticipate generating up to 2x 94 = 188 lines within the current OREGIN funding. We are interested in extending this in discussion and collaboration with partners worldwide.
- For B. rapa, a DFFS is currently being compiled by Guusje Bonnema and colleagues. More details will be made available here as work progresses.